Terms classification for every Exponential Random Graph Model (ERGM)
Terms can be classified in six main ways.
Dyadic independent and dyadic dependent terms: We encounter the first one when the probability of edge formation is related to nodes properties or attributes; we encounter the second when the probability of edge formation depends on other existing edges.
Structural and nodal attributes terms: The first kind provides tools to understand the structure of the network per se; the second kind provides tools to explain how nodal attributes might have influenced the formation of edges.
Terms for directed networks and terms for undirected networks
Exogenous and Endogenous terms: The first one refers to terms using covariates, the second to structural terms.
Markovian or non-Markovian: a Markovian term measures the structure in a network neighborhood
Curved (geometrically weighted ) or non-curved: terms that are tweaked to improve the model stability
Binary Exponential Random Graph Model (ERGM)
An ERGM model is performed through the ergm::ergm
function. The basic function call is as follows:
fit <- ergm::ergm(formula)The formula requires the specification of a network dependent variable, and a list of terms.
Most popular structural/endogenous - dyadic independent terms
edgesExtent to which the number of edges in the network characterizes the overall structure (Is it a random number of edges, or it is the meaningful outcome of a certain phenomenon?). Introduces one statistic to the model. Directed and Undirected networks.densityExtent to which the network density characterizes the overall structure (Is it a random density, or it is the meaningful outcome of a certain phenomenon?). Introduces one statistic to the model. Directed and Undirected networks.senderExtent to which a specific node, compared to a baseline one, is sending out non-random edges (different from the same node’s behavior in a random distribution). Introduces to the model as many statistics as the number of nodes minus one. Directed Networks only.receiverExtent to which a specific node, compared to a baseline one, is receiving non-random edges (different from the same node’s behavior in a random distribution). Introduces to the model as many statistics as the number of nodes minus one. Directed Networks only.
Most popular structural/endogenous/Markovian - dyadic dependent terms
mutualExtent to which ties are more likely to be reciprocated than they would be in a random network (controlling for the other effects). Introduces one statistic to the model. Directed networks only.asymmetricExtent to which the observed non reciprocated ties are non-random. Introduces one statistic to the model. Directed networks only.trianglesExtent to which the observed triangles are non-random. Introduces one statistic to the model. Directed and Undirected networks. In the case of directed network measures “transitive triple” and “cyclic triple”, so triangle equals tottripleplusctriple.triadcensusExtent to which the sixteen categories in the categorization of Davis and Leinhardt (1972) are observed in the network and are not generated at random. Introduces 16 statistics to the model. Directed networks only.
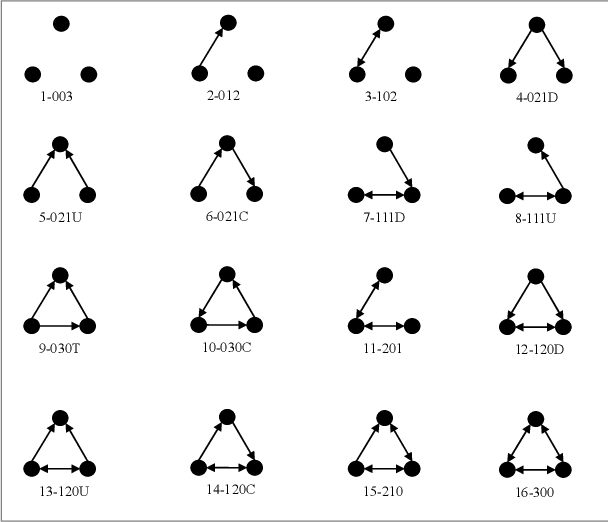
balanceExtent to which type 102 or 300 in the categorization of Davis and Leinhardt (1972) -balanced triads- observed in the network are non-random. Introduces one statistic to the model. Directed networks only.transitiveExtent to which type 120D, 030T, 120U, or 300 in the categorization of Davis and Leinhardt (1972) -transitive triads- observed in the network are non-random. Introduces one statistic to the model. Directed networks only.intransitiveExtent to which type 111D, 201, 111U, 021C, or 030C in the categorization of Davis and Leinhardt (1972) -intransitive triads- observed in the network are non-random. Introduces one statistic to the model. Directed networks only.degree(n),idegree(n),odegree(n)Extent to which nodes with a specified degree are non random. Introduces one statistic to the model. Directed and Undirected networks, with the possibility ofinandoutspecifications for Directed networks.kstar(n),istar(n),ostar(n)Extent to which stars connecting the specified number of nodes are non random. Introduces one statistic to the model. Directed and Undirected networks, with the possibility ofinandoutspecifications for Directed networks.cycle(n)Extent to which cycles with a specified number of nodes are non-random. Introduces one statistic to the model. Directed and Undirected networks.
Most popular structural/endogenous/curved - dyadic dependent terms
gwesp(decay=0.25, fixed=FALSE)Geometrically weighted edgewise shared partner distribution. It can be used in place of triangles to improve convergence. The decay parameter should be non-negative. The value supplied for this parameter may be fixed (iffixed=TRUE), or it may be used instead as the starting value for the estimation of decay in a curved exponential family model (whenfixed=FALSE, the default) (see Hunter and Handcock, 2006). This term can be used with directed and undirected networks. For directed networks, only outgoing two-path (“OTP”) shared partners are counted.dgwesp(decay=0.25, fixed=FALSE, type= 'RTP')Geometrically weighted edgewise shared partner distribution. It also counts other types of shared partners not covered bygwesp: Outgoing Two-path (“OTP”), Incoming Two-path (“ITP”), Reciprocated Two-path (“RTP”), Outgoing Shared Partner (“OSP”), Incoming Shared Partner (“ISP”).gwdegree(decay, fixed=FALSE, attr=NULL, cutoff=30, levels=NULL),gwidegree(.5,fixed=T),gwodegree(.5,fixed=T)Geometrically weighted degree distribution. It can be used in place ofdegree(n)to improve convergence. Introduces one statistic to the model equal to the weighted degree distribution with decay controlled by the decay parameter. Directed and Undirected networks, with the possibility ofinandoutspecifications for Directed networks.
Most popular nodal covariate terms
nodecov,nodeicov,nodeocovNumeric or Integer attributes. Extent to which the attribute values influence edge formation (same as in a logit model) so that it is non-random under that condition. Introduces one statistic to the model. Directed and Undirected networks, with the possibility ofinandoutspecifications for Directed networks. Dyadic independent.nodefactor,nodeifactor,nodeofactorCategorical attributes. Extent to which nodes characterized by a specific category form more ties, so that tie formation is non-random under that condition. Introduces to the model a number of statistics equal to the number of categories minus one. Directed and Undirected networks, with the possibility ofinandoutspecifications for Directed networks. Dyadic independent.absdiffNumeric or Integer attributes. Extent to which common features measured in terms of distance similarity influence edge formation, so that edge formation is non-random under that condition. Introduces one statistic to the model. Directed and Undirected networks. Dyadic independent.nodematchCategorical attributes. Extent to which nodes characterized by a specific category belonging to a certain attribute form ties with other node characterized by the same category, so that tie formation under that condition is non-random. Introduces to the model as many statistics as the number of categories. Directed and Undirected networks. Dyadic independent. —Differential homophilyedgecovMatrix attribute. Extent to which the ties formed in another context influence tie formation in the context of the current model, so that tie formation under that circumstances is non-random. Introduces one statistic to the model. Directed and Undirected networks. Dyadic dependent.nodemixCategorical attributes. Extent to which nodes denoted by different categories of an attribute form ties, so that tie formation under these circumstances is non-random. Introduces as many statistics as the number of combinations between every two categories. Directed and Undirected networks. Dyadic independent.
Terms specifications
Use the argument levels within the term specification
for selecting the baseline or reference category.
Example: set female as a reference category.
fit <- ergm::ergm(Net ~ edges + nodefactor('sex', levels = -(2)))Searching for terms
You can look for additional terms with
search.ergmTerms(keyword, net, categories, name)You have four arguments to help you finding terms:
keywordoptional character keyword to search for in the text of the term descriptions. Only matching terms will be returned. Matching is case insensitive.neta network object that the term would be applied to, used as template to determine directedness, bipartite, etccategoriesoptional character vector of category tags to use to restrict the results (i.e. ‘curved’, ‘triad-related’) –see categorization of terms in the manualnameoptional character name of a specific term to return
Checking your data before the analysis
Before you run any exponential random graph model you must know your data by heart. Not only using descriptive network statistics, but also checking model specifications, before hitting the run button.
- Manually check the attribute(s) (numeric, integer, categorical, ordinal)
table(snafun::extract_vertex_attribute(Net, 'sex'))- check mixing of categorical attributes
snafun::make_mixingmatrix(Net, "sex")- check model statistics.
summary(Net ~ edges + nodefactor('sex'))This last one provides the number of observed cases under the assumptions of each term.
Reading results
You interpret ERGM results as logit models results. Two options:
- Compute odd ratios for each coefficient
OR <- exp(coef)- Compute probability for each coefficient
- Compute odd ratios using the
SNA4DSfunction
OR <- snafun::stat_ef_int(m)- Compute probability using the
snafunfunction
P <- snafun::stat_ef_int(m, type = "probs")Simulating networks
It is sometimes helpful to simulate networks with the same features at the one you observed in real life.
- Simulating a network from a model
- simulate network fixing the coefficient results
RandomNet <- network::network(16,density=0.1,directed=FALSE)
sim <- simulate(~ edges + kstar(2), nsim = 2, coef = c(-1.8, 0.03),
basis = RandomNet,
control = ergm::control.simulate(
MCMC.burnin=1000,
MCMC.interval=100))
sim[[1]]MCMC Diagostics
You can check the Monte Carlo Markov Chains diagnostic for your dyadic dependent model using the function:
ergm::mcmc.diagnostics(fit)Goodness of Fit
You can check the goodness of fit of your model using the function
ergm::gof(fit)You can also plot your gof output
or, making use of your new best friend (the snafun
package):
snafun::stat_plot_gof(fit)
stat_plot_gof_as_btergm(fit)Bipartite ERGMs
A bipartite ERGM works exactly the same way as a binary one. However, in order to make it handle data differentiating between two partitions, it is necessary to use some specially defined terms. Moreover, you will need to specify the model with more advanced settings since it is computationally more demanding.
Importing Bipartite ERGMs
Since you are using the same function as binary ERGM
(ergm::ergm) to run the model, it is necessary to make sure
that the software knows that it needs to handle a bipartite
structure.
Step one: Specify the incidence matrix
The data that contains bipartite network information needs to be specified into a partition 1 X partition 2 data frame or matrix.
If 10 people attend 4 events, the incidence matrix will have dimensions 10 X 4.
Step two: Import the network as a bipartite
You can import the network as bipartite using these specifications.
BipNet <- network::network(BipData, directed = FALSE, bipartite = TRUE)Step three: Import the attributes
You can import the attributes using this code. The attribute vector needs to contain as many elements as Partition 1 + Partition 2. However, it is unlikely to have an attribute that makes sense for both partitions at the same time.
Make sure to insert the information that concerns partition 1 and afterward the information that concerns partition 2.
Make sure to code as NA the entry for the partition for
which you do not have information.
For instance, if we have one attribute for partition one in a network with 10 nodes in partition 1 and 4 in partition 2, we will have the first ten digits storing information about nodes in partition 1 and 4 NAs for partition 2.
# vertex.names <- vector of names
attrib1 <- as.character(c(0, 0, 0, 0, 0, 1, 1, 1, 1, 1, NA, NA, NA, NA))
snafun::add_vertex_attributes(BipNet, "vertex.names", vertex.names)
snafun::add_vertex_attributes(BipNet, "attrib1", attrib1)Step four: Bipartite extra info
In order to make sure that the software correctly reads your bipartite network, you need to code an extra attribute.
The attribute focuses on partition one. For example, if we have 10 nodes in partition 1 we will use:
snafun::add_vertex_attributes(BipNet, "bipratite", value = rep(10, 10), v = 1:10)Note: bipratite, yes, it is a typo, but a typo in the
package. Hence make sure you misspell it; otherwise, you will get an
error. (FUN FACT!)
Terms for Bipartite ERGMs
Bipartite ERGMs terms are provided at the same time for both partitions since it is relevant to consider the same structure from both perspectives.
You can find the full list of bipartite terms by running:
ergm::search.ergmTerms(categories = "bipartite")They are 32 in total, so it is manageable.
The most popular terms are:
b1star(k)&b2star(k)–star(k)for binary ERGMsgwb1dsp()&gwb2dsp()– same asgwdsp()for binary ERGMsb1cov&b2cov– same asnodecov()for binary ERGMsb1factor&b2factor– same asnodefactor()for binary ERGMsb1nodematch&b2nodematch– same asnodematch()for binary ERGMs
Specifying advanced options
Since a bipartite ERGM is computationally more demanding than a
regular one, you need to make sure you specify the advanced options
offered by the ergm package.
Constraints
Constraints are options that allow you to set limits your simulation takes into account
For instance, you can limit the simulation setting a min and a max degree.
constraints= ~ bd(minout = 0, maxout = 7)
For instance,
m <- ergm::ergm(BipNet ~ edges + b1factor("attr1", levels = -1) + b1star(2),
constraints= ~ bd(minout = 0, maxout = 7))There are many other constraints that it is possible to use
Control
Controls are options that allow you to be aware of what your simulation is doing to a larger extent and, for this reason, to make it faster.
There are several options. For instance,
MCMC.burnin- ignore than many chains before starting to estimate parametersMCMC.samplesize- collect that number of information from the previous state in order to inform the following oneseed- makes the simulation go the same way every time it is runMCMLE.maxit- breaks the algorithm after that number of attempts.
For instance,
m <- ergm::ergm(BipNet ~ edges + b1factor("attr1", levels = -1) + b1star(2),
constraints= ~ bd(minout = 0, maxout = 7),
control = ergm::control.ergm(MCMC.burnin = 5000,
MCMC.samplesize = 10000,
seed = 1234,
MCMLE.maxit = 20))Weighted ERGMs
A weighted network is a network where the edges express the weight or the intensity of the relationship.
It is still unimodal, but it contains more information.
In order to use weighted ergms, the network needs to be fully connected.
Different kinds of weights require different models. You need to check:
- what kind of variable type characterizes your weights (integer, count, numeric, ordinal…)
- what kind of distribution does your weight have
The GERGM package works with fully connected networks
and (theoretically ) every kind of weight variable.
The GERGM package does not recognize either the
network or the igraph classes.
You need to work with weighted adjacency matrices (NxN - squared, that has inside the weight, rather than 0s and 1s).
Documentation
Installation:
remotes::install_github("matthewjdenny/GERGM", dependencies = TRUE)
User manual
https://github.com/matthewjdenny/GERGM
Vignette
Run browseVignettes("GERGM")in the R console
Most popular endogenous terms (Markovian and Curved)
twostars, equivalent tostar(2)inergmout2stars, equivalent toostar(2)inergmin2stars, equivalent toistar(2)inergmctriads, same as inergmmutual, same as inergmttriads, same as inergm
Convergence problems? Use exponential down-weighting. AKA “curve” the terms:
E.g., out2stars(alpha = 0.25) - default = 1
Most popular exgenous terms
absdiff(covariate = "MyCov")sender(covariate = "MyCov")– different fromergm, here you can insert an attributereceiver(covariate = "MyCov")– different fromergm, here you can insert an attributenodematch(covariate = "MyCov", base = "Ref.cat")nodemix(covariate = "MyCov", base = "Ref.cat")netcov(network)– likeedgecovinergm
Running the model
First, we specify the formula,
formula <- adjacencyMatrix ~ edges +
sender("myCov") +
receiver("myCov") +
netcov(otherAdjMat) +
mutual(alpha = .9)Then we run the model
set.seed(5)
gergmResults <- GERGM::gergm(formula,
estimation_method = "Metropolis", # chose the algorithm to estimate the model
covariate_data = covariateData, # passing attributes on
number_of_networks_to_simulate = 100000, # same as ergm
MCMC_burnin = 10000, # same as ergm
thin = 1/10, # retaining only a small number of simulated Networks in the computer memory
transformation_type = "Cauchy") # distribution of the weightThe GERGM::gergm function automatically prints results
while it runs. However, it is also possible to print them again
separately.
Printing a table with standard errors and coefficients
(EstSE <- rbind(t(attributes(gergmResults)$theta.coef),
t(attributes(gergmResults)$lambda.coef)))Significance
GERGM models use confidence intervals instead of
p-values.
You can estimate the confidence interval using this formula
If the lower and the upper intervals are both negative or both positive, the coefficient is significant.
If the lower and the upper intervals have different sign, the coefficient is not significant.