Graph level indices
Below you will find a table to determine the foundational indices of a graph at the graph-level.
| Explore the graph | |
| Summarize the network | |
|---|---|
| snafun | |
| igraph | — |
| network | — |
| Count the number of vertices | |
| snafun | |
| igraph | |
| network | |
| Count the number of edges | |
| snafun | |
| igraph | |
| network | |
| Density | |
| snafun | |
| igraph | |
| network | |
| Reciprocity | |
| snafun | |
| igraph | |
| network | |
| Transitivity | |
| snafun | |
| igraph | |
| network | |
| Mean distance between vertices | |
| snafun | |
| igraph | |
| network | — |
| Degree distribution | |
| snafun | |
| igraph | |
| network | — |
| Dyad census | |
| snafun | |
| igraph | |
| network | |
| Triad census | |
| snafun | |
| igraph | |
| network | |
| Degree assortativity | |
| snafun | — |
| igraph | |
| network | — |
| Diameter | |
| snafun | |
| igraph | |
| network | — |
| Radius | |
| snafun | |
| igraph | |
| network | — |
| Compactness | |
| snafun | |
| igraph | — |
| network | — |
| Centralization | |
| snafun | |
| igraph1,2 | |
| network1 | |
| Mixing matrix | |
| snafun | |
| igraph | — |
| network | |
| Correlation between two graphs | |
| snafun | |
| igraph | — |
| network | |
| Fast-greedy community detection | |
| snafun | |
| igraph | |
| network | — |
| Girvan-Newman community detection | |
| snafun | |
| igraph | |
| network | — |
| Louvain community detection | |
| snafun | |
| igraph | |
| network | — |
| Walktrap community detection | |
| snafun | |
| igraph | |
| network | — |
| Merge community membership | |
| snafun | |
| igraph | — |
| network | — |
| 1 igraph and sna only calculate `Freeman` centralization (snafun does both `Freeman` and `sd`) | |
| 2 `$res` or `$vector` return the centrality scores | |
Communities and other subgroups
It is informative to know that the snafun functions to
extract communities yield results that can be scrutinized by
igraph. The snafun package is smart enough to
do this, regardless of whether the original input graph was of class
igraph or network. Really handy.
Here’s an example.
# generate a random directed graph with 20 vertices and 30 edges
g <- snafun::create_random_graph(20, "gnm", m = 30)
# determine the walktrap communities
walk <- snafun::extract_comm_walktrap(g)
print(walk)
#> IGRAPH clustering walktrap, groups: 6, mod: 0.33
#> + groups:
#> $`1`
#> [1] 7 9 10 12 13 18
#>
#> $`2`
#> [1] 5 6 16 19 20
#>
#> $`3`
#> [1] 1 2 14
#>
#> $`4`
#> + ... omitted several groups/vertices
# get the modularity score
igraph::modularity(walk)
#> [1] 0.3344444
# who is member of which community
igraph::communities(walk)
#> $`1`
#> [1] 7 9 10 12 13 18
#>
#> $`2`
#> [1] 5 6 16 19 20
#>
#> $`3`
#> [1] 1 2 14
#>
#> $`4`
#> [1] 3 15 17
#>
#> $`5`
#> [1] 4 11
#>
#> $`6`
#> [1] 8
# which community is a vertex member of
igraph::membership(walk)
#> [1] 3 3 4 5 2 2 1 6 1 1 5 1 1 3 4 2 4 1 2 2
# number of communities
length(walk)
#> [1] 6
# size of each community
igraph::sizes(walk)
#> Community sizes
#> 1 2 3 4 5 6
#> 6 5 3 3 2 1
# which edge connects multiple communities
igraph::crossing(walk, g)
#> [1] FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [13] FALSE FALSE TRUE TRUE TRUE TRUE FALSE FALSE TRUE FALSE TRUE TRUE
#> [25] TRUE TRUE TRUE FALSE TRUE FALSE
# plot the network, highlighting the communities
plot(walk, g)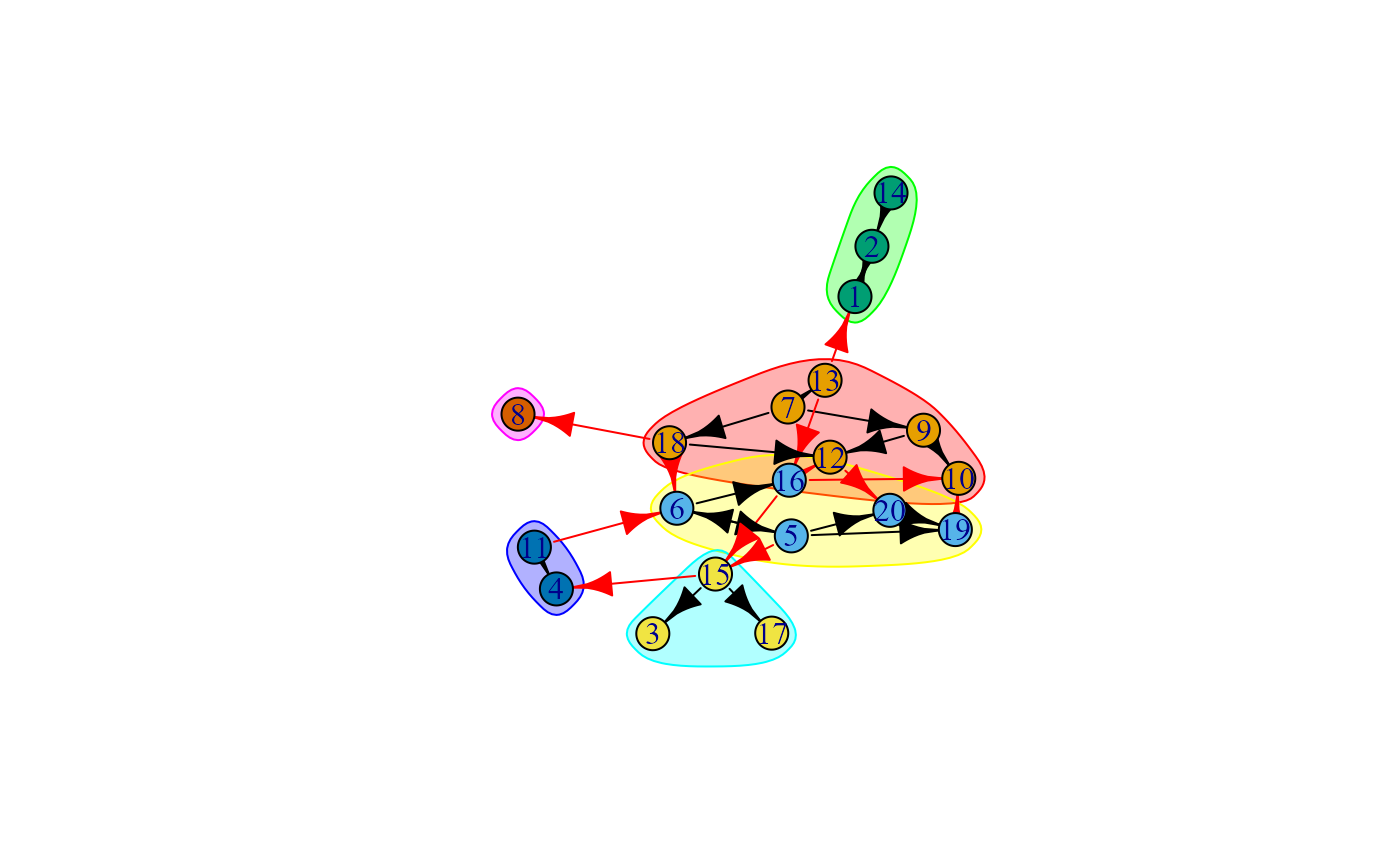
If you are so inclined, you can plot the community division as a dendrogram, as follows:
snafun::plot_comm_dendrogram(walk)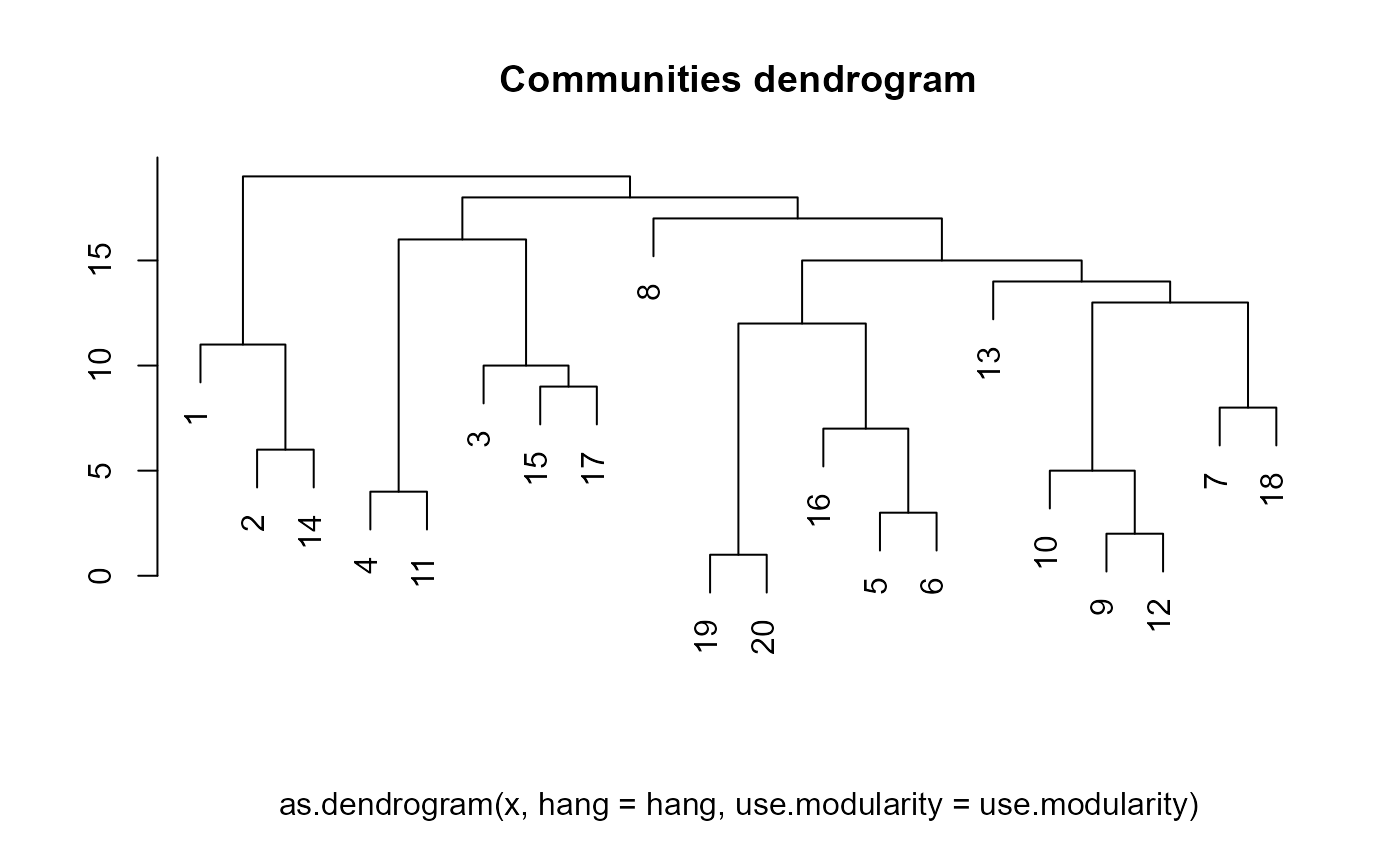
Vertex-level indices
Here are the functions to determine many of the vertex-level indices you will want to use in this course.
| Explore the vertices | |
| Degree | |
|---|---|
| snafun | |
| igraph | |
| network | |
| Betweenness | |
| snafun | |
| igraph | |
| network | |
| Flow betweenness | |
| snafun | — |
| igraph | — |
| network | |
| Bonacich power centrality | |
| snafun | — |
| igraph | |
| network | |
| Closeness centrality | |
| snafun | |
| igraph | |
| network | |
| Harmonic centrality | |
| snafun | |
| igraph | |
| network | — |
| Stress centrality | |
| snafun | |
| igraph | — |
| network | |
| Eccentricity | |
| snafun | |
| igraph | |
| network | — |
| Eigenvector | |
| snafun | |
| igraph | |
| network | |
| Page rank | |
| snafun | |
| igraph | |
| network | — |
| Geo k-path | |
| snafun | |
| igraph | — |
| network | — |
| Shapley centrality | |
| snafun | |
| igraph | — |
| network | — |
| Who are the neighbors of a vertex | |
| snafun | |
| igraph | |
| network | |
| Neighborhood of a vertex1 | |
| snafun | — |
| igraph | |
| network | |
| 1 These functions serve equivalent purposes, but yield quite different kinds of outputs | |